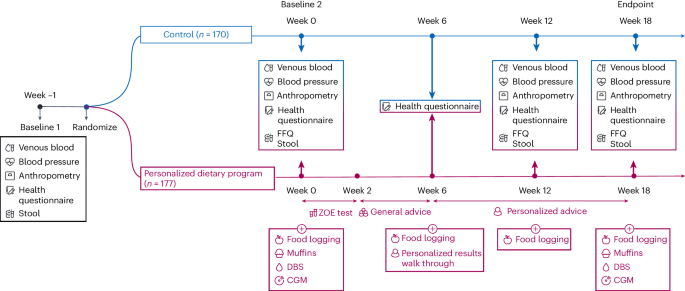
Study design
The ZOE METHOD study was an 18-week parallel-design, randomized controlled trial. The trial was registered on ClinicalTrials.gov (ClinicalTrials.gov registration: NCT05273268) and listed as the ZOE METHOD Study: Comparing Personalized versus Generalized Nutrition Guidelines. The remote trial carried out in the US compared standard care dietary advice (control) versus a PDP in a cohort generally representative of the US adult population. Standard care dietary advice (United States Dietary Guidelines for Americans, 2020–2025) was delivered in the form of an USDA dietary recommendations digital leaflet, a short video lesson, access to online resources and regular check-ins. The PDP provided dietary advice using the ZOE 2022 algorithm, incorporating food characteristics, individuals’ glucose control and postprandial TG concentrations3, individuals’ microbiomes13, atherosclerotic cardiovascular disease risk and health history, to produce personalized food scores delivered during an 18-week program alongside more generalized nutrition and lifestyle education through a remote mobile phone application (the ZOE app). Ethical approval for the trial was obtained through the Advarra IRB (IRB no. 00000971; protocol no. 00044316). All participants provided written informed consent and the study was carried out in accordance with good clinical practice and the Declaration of Helsinki (2013). Outcome measurements were made at baseline and after randomization to their respective treatments.
Participant selection and randomization
Males and females reflective of the average US adult population (aged 40–70 years; waist circumference greater than ethnicity-specific and sex-specific 25th percentile values; fruit and vegetable intake below 450 g per day (to capture 75% of the population)) living in the US were recruited (1 March 2022 to 10 August 2022) by electronic advertisement (e-mail to the Stanford Nutrition Studies Research Cohort, the Empowered Gut newsletter and the ZOE Ltd mailing lists). Both sexes were eligible for recruitment and sex was determined using self-reported questionnaires with the following question: ‘What sex were you assigned at birth?’ Through the recruitment channels (e-mail and website), participants were invited to complete an online screening questionnaire and then invited to attend a primary baseline clinical visit (described in detail below) where all eligibility criteria were assessed. After this two-step screening process, participant eligibility was confirmed and a minimization-randomization program (MinimPy v.0.3, Python Package Index; pypi.org/project/MinimPy/) was used for treatment allocation. Participants were randomly and equally allocated to one of the two treatments based on the following minimization factors: (1) sex, male or female; (2) waist circumference, above or below their ethnicity-specific median; and (3) fruit and vegetable intake, above or below the median US adult intake of 234 g per day. Trained study coordinators enrolled, assigned and informed participants about their allocation to treatment via e-mail. Participants were informed of all study procedures before providing electronic consent. Participants were excluded from the study if any of the following criteria applied: had taken part in the ZOE product or any PREDICT study beforehand; were unable to read and write in English, as the ZOE app is only available in English; did not complete the first Quest visit successfully; had an iOS/Android device not compatible with the app; used medications affecting lipids (lipid-lowering drugs, for example, statins; antidiabetic medications, for example, metformin and insulin), and supplements including fish oil (unless willing to safely come off these for 4 weeks before the start of the study, and for the duration of study); had ongoing inflammatory disease, for example, rheumatoid arthritis, systemic lupus erythematosus, polymyalgia and other connective tissue diseases; had cancer in the last 3 years, excluding skin cancer; had chronic gastrointestinal disorders, including inflammatory bowel disease or celiac disease (gluten allergy), but not including irritable bowel syndrome; were taking the following daily medications: immunosuppressants, corticosteroids or antibiotics in the last 3 months, not including inhalers; were users of prescription proton pump inhibitors, such as omeprazole and pantoprazol, unless they were able to stop 2 weeks before the start of the study and remained off them for the entire duration of the study (provided their treating physician deemed it safe for them to do so); were currently suffering from acute clinically diagnosed depression or anxiety disorder; had a heart attack (myocardial infarction) or stroke in the last 6 months; were pregnant or planning pregnancy in next 12 months, or were breastfeeding; were vegan, had an eating disorder or were unwilling to take foods that were part of the study; had an allergy to adhesives, which would prevent proper attachment of the CGM.
Interventions and procedures
The study design is summarized in Fig. 1.
Primary baseline testing (week −1)
Baseline clinical visit
Participants attended a baseline clinical visit at the Quest Diagnostic Patient Service Center, where baseline measures were assessed, including a fasted venous blood draw, and anthropometric measurement of height, body weight, hip circumference, waist circumference and blood pressure. Participants who did not attend a clinic visit within 1 week of their visit date were withdrawn from the study.
Health questionnaire
Participants remotely completed two questionnaires administered through an online survey before randomization. These questionnaires included (1) a primary questionnaire capturing baseline health status and medical health history and (2) a secondary questionnaire capturing information on anthropometrics, sleep, energy level, mood, hunger, skin, female health (menopause) and current medication use.
Participant survey
A survey where participants confirmed completion of the primary baseline study tasks was administered at the end of week −1 to assess participant compliance.
Stool sample collection
Stool samples for microbiome analysis (required for the algorithm predictions) were collected by participants at home using the DNA/RNA SheildTM Fecal Collection Tube (Zymo Research) containing buffer (catalog no. R1101, Zymo Research). Once collected, the sample was stored at room temperature before being shipped to the analyzing laboratory inside a prepaid return kit.
Secondary baseline testing (week 0)
Baseline measures
After allocation to treatment, both PDP and control groups completed a secondary set of baseline measurements, including fasted venous blood tests, questionnaires and stool collection as described in the primary baseline testing section. Approximately 1 week after their primary clinical visit, participants completed a secondary visit to the Quest center. Non-completion of this second visit within the required time period resulted in participant withdrawal from the study. In addition to this, participants completed an FFQ. The PREDICT FFQ, which captured information on 264 foods, food groups and beverages over the previous month was administered via an online survey3. For the control group, links to the FFQ were provided via e-mail. For the PDP group, links to the FFQ were provided via e-mail or via the ZOE app.
ZOE test kit
PDP participants were additionally asked to complete the ZOE test kit. This included (1) a CGM, (2) standardized test meals (three muffins) and (3) a DBS. Participants applied and wore a CGM (Freestyle Libre 2, Abbott) on their upper arm for up to 14 days. Two days after CGM application, participants completed 2 days of standardized meal intervention. Meals consisted of muffins with mixed macronutrient composition and were consumed for breakfast and lunch (day 1, as a sequential mixed meal intervention) and for breakfast only (day 2). Breakfast meals were consumed after an overnight fast of at least 8 h. Participants were asked to consume the entire portion of the meal provided within 15 min. The consumption of their meal was scanned in the app using the unique barcode labeled on each meal. The time participants started and completed eating their meal was recorded. They were asked to report any deviations from this protocol to study staff.
After the sequential test meal, a finger-prick DBS test was completed (6 h after breakfast to measure postprandial responses). Blood test cards were stored at room temperature until shipping to the analyzing laboratory via a prepaid return mailing kit. Finally, after completion of their test meals, participants were asked to log their habitual diet through the ZOE app. This app provided the functionality of a weighed food diary as well as a log of all the study tasks required of the participant during the ZOE test kit phase.
Dietary advice
Participants in the control group were e-mailed a PDF file containing a digital leaflet from the USDA Dietary Guidelines for Americans (2020–2025) accompanied by a video verbalizing the dietary advice, in accordance with a typical general consultation. In addition, participants were provided with online resources. Study coaches were available by e-mail to answer questions and provide support. The USDA guidelines recommend daily or weekly amounts from different food groups to maintain a healthy lifestyle. Participants were advised to follow this dietary advice for the study duration (weeks 2–18). Each week they received an e-mail from a study nutrition coach to check in.
PDP participants received generalized nutrition and lifestyle advice through the ZOE app, which they followed for 4 weeks (weeks 2–6), while personalized results were being generated via the ZOE 2022 algorithm (see Fig. 1 for more details). Generalized advice was presented via the app in the form of interactive ‘lessons’ as part of a program of learning. The lessons covered basic nutritional and dietary health concepts, including dietary diversification, increasing plant food consumption, increasing fiber intake, replacing refined carbohydrates with wholegrains and consumption of fermented foods.
At week 6, PDP participants received a personalized ‘Insights’ study report, including a personalized blood sugar score, blood fat score, gut diversity score, gut microbiome score and presence or absence of several microbial species2. These reports also included results from the ZOE 2022 algorithm, specifically information about person-specific food scores.
The interventions were not matched for contact or intensity to test the efficacy of the PDP, which involves personalized diet scores overlaid with generalized dietary and lifestyle advice delivered as a set of program lessons.
Personalized food quality scores
A personalized ZOE food quality score was computed using the ZOE 2022 algorithm for each food item consumed by the PDP participants. Food quality scores were based on both the macronutrients of a food item and further food metadata, including glycemic load, fat quality, level of processing and food group (for example meat, fruit, vegetables and fermented foods). They were personalized to an individual’s glucose control, postprandial TG concentration, atherosclerotic cardiovascular disease risk, health history and microbiome composition (abundance of specific health-promoting and health-reducing microbial taxa and the associations of these taxa with food items). The ZOE 2022 algorithm was trained using expert input on appropriate food quality scores for different individual phenotypes for a small number of foundational foods, and was used to predict personalized food quality scores for all individual phenotypes and all food items, which were then further personalized for detailed microbiome composition.
The food quality scores ranged from 0 to 100, with higher values indicating more healthful meals. Based on this food quality score, personalized recommendations could be made, that is, consume foods with a quality score of 0–24 once in a while, enjoy in moderation foods with a score of 25–49, enjoy foods regularly with a score of 50–74 and enjoy foods freely with a score of 75–100. A participant’s personalized meal scores throughout the day were combined by further algorithms to generate personalized day scores also ranging from 0 to 100. Throughout the study, participants were instructed to consume a diet (and record it in the app) reaching a certain day score threshold, which increased throughout the study duration, to the best of their ability. These day scores were accessible to participants, aiming to motivate them and convey to them their compliance to their dietary advice. The diet did not involve calorie restriction or calorie counting.
From week 6, PDP participants received personalized food scores and meal recommendations within the ZOE app. PDP participants were asked to attend a single phone or video call with a study staff member to discuss their results and to make these results immediately accessible to and actionable by the participant. Following this, a set of program lessons was administered in the app for 12 weeks (termed the ‘action plan’) during which participants were taught how to engage with and adhere to their personalized plan. Contact with study coaches was available via the app.
Week 12 measures
PDP and control groups completed a set of measures at week 12, including fasted venous blood tests (Quest visit), questionnaires, stool collection and FFQ as described in the primary and secondary baseline testing sections above.
Endpoint measures (week 18)
Endpoint data collection was completed in the 19th week of the study, at which point both groups had been allocated to their respective treatments for 18 weeks. PDP and control groups completed a set of endpoint measures, including fasted venous blood tests (Quest visit), questionnaires, stool collection and FFQ as described in the primary and secondary baseline testing sections above.
PDP participants were provided a second ZOE test kit to retest their nutritional responses, including application of a second CGM, consumption of the standardized meal intervention and completion of DBS.
Additional follow-ups
PDP participants were followed up at 8 and 12 months with a clinical visit, including fasted venous blood tests, questionnaires, stool collection and FFQ as described in the primary and secondary baseline testing sections above. Control participants were given the option to join a nested cross-over arm on completion of the 18-week endpoint measures. These participants completed the PDP arm protocol and completed the 6-, 12- and 18-week measures. Alternatively, control participants were offered the ZOE nutrition commercial product.
Participants were recruited from March 2022 to August 2022. The core intervention period took place from April 2022 to February 2023, and follow-ups were completed by September 2023.
Adherence
As part of the study design, participants in both arms were asked to self-report adherence (scale 0–10) to the dietary advice given by the questionnaire administered every 6 weeks (week 7, week 12 and week 18 for the control group; week 12 and week 18 for the PDP group) during the study period. As part of the PDP only, participants were asked to record their dietary intake in real time on a minimum of four consecutive days (including one weekend day and 1,200 kcal or more per day) per month using a designated smartphone app (ZOE app). Each food item was recorded along with weight or portion units by selecting the food from a database (the USDA compositional database and a commercial database) containing approximately 900,000 items. Adherence to the PDP was evaluated through logging metrics and self-recorded dietary intake in the logging app.
Outcomes
Specified primary outcomes were serum TG concentration and direct LDL-C concentration. The primary outcome was the 18-week change from baseline. Therefore, secondary outcomes were changes in weight, waist circumference, hip circumference, systolic blood pressure and diastolic blood pressure, blood HbA1c, serum insulin, serum glucose, serum C-peptide, serum apolipoprotein A1, serum apolipoprotein B, fecal gut microbiome (species richness, Shannon diversity and Bray–Curtis dissimilarity), postprandial blood TG concentration, habitual diet quality (HEI) and self-reported energy level. Other outcomes included self-reported mood, hunger, total protein, albumin, globulin, bilirubin, alkaline phosphatase, aspartate aminotransferase, alanine aminotransferase, C-reactive protein, tumor necrosis factor alpha and full blood count.
DBS collection and processing
Postprandial TG (mmol l−1), high-density lipoprotein cholesterol (mmol l−1) and cholesterol (mmol l−1) were quantified from finger-prick DBS (Clinical Reference Laboratory) tests completed by PDP participants in weeks 0 and 18 of the study (during completion of the ZOE test kit). DBS tests were completed 360 min after consuming the breakfast test meal. After washing their hands, participants pricked a finger with a sterile lancet and placed 3–4 drops of blood on their test card. Study staff assessed test validity using a photo and time point of testing logged by the participant in the app. Test cards not meeting the quality protocol (multiple small spots or inadequate coverage) were not included in the analysis. Participants were encouraged to complete the sequential test meal and DBS test again when either of these was inadequately completed. Each test card was stored in a foil pouch with a desiccant packet once completed and mailed to the analyzing laboratory in a prepaid kit within 24 h of completion.
Analysis was done at the Clinical Reference Laboratory. Advance Dx100 Technology DBS cards were analyzed for lipemic metabolites by the Clinical Reference Laboratory. Portions of test cards were taken from the sample, from which the dried blood was extracted and analyzed using standard quantification methods.
Fasted venous blood collection and processing
Fasted venous blood draws were performed at Quest Diagnostic Patient Service Centers and processed by Quest Diagnostics; 500 μl of venous blood was collected in serum separator tubes (SSTs). Then, 250 μl of venous blood was collected in EDTA tubes. SSTs and EDTA tubes were left at room temperature for 30 min (or up to 1 h) and centrifuged at 1,600g for 15 min at 4 °C. Direct LDL-C, TG, glucose, insulin, C-peptide, apolipoprotein A1 and apolipoprotein B were quantified in serum (SST), and HbA1c was quantified in whole blood (EDTA). The full list of clinical blood chemistry measures quantified in this study are shown in Supplementary Table 10.
Continuous glucose monitoring
Interstitial glucose was measured every minute and aggregated into 15-min readings, using the Freestyle Libre 14-day CGM (Abbott Diabetes Care). Participants randomized to the PDP group were instructed to apply the CGM two days before starting their standardized meal intervention, to the upper, nondominant arm and to cover the monitor with an adhesive patch (Sourceful) for improved durability. CGMs were worn for up to 14 days and participants were unblinded to the results. Given that the CGM device requires time to calibrate once applied, CGM data collected 12 h and onwards after activating the device was used for the analysis.
Fecal sampling and microbiome testing
DNA extraction and sequencing
On receipt in the laboratory, samples were homogenized, aliquoted and stored at −80 °C in QIAGEN PowerBeads 1.5-ml tubes and used to extract bacterial DNA. All 815 stool samples were processed and analyzed using a Shotgun Metagenomic Sequencing Service (Zymo Research). The DNA was first isolated using the ZymoBIOMICS 96 MagBead DNA Kit (Zymo Research). Then, the sequencing libraries were prepared using the Illumina DNA Library Prep Kit with up to 500 ng DNA input according to the manufacturer’s protocol, using unique dual-index 10-bp barcodes with Nextera adapters (Illumina). The libraries were pooled in equal abundance and the final pools were quantified using quantitative PCR and a TapeStation (Agilent Technologies). The final libraries were sequenced using the NovaSeq 6000 platform (Illumina) according to the manufacturer’s protocols, generating 150-bp paired-end reads. The NovaSeq control software NCS v.1.5 was used. Image analysis, base calling and quality checking were performed with the Illumina data analysis pipeline RTA3.3.5 and bcl2fastq v.2.20.
Metagenome quality control and preprocessing
All sequenced metagenomes were preprocessed using the pipeline implemented in github.com/SegataLab/preprocessing. Briefly, the pipeline consisted of three steps: the first step involved read-level quality control and removed low-quality reads (Q 45) with the ‘–sensitive-local’ parameter, allowing confident removal of the phi X 174 Illumina spike-in and human-associated reads (hg19 reference human genome release). The last step consisted of splitting and sorting the cleaned reads to create standard forward, reverse and unpaired read output files for each metagenome (average: 35 ± 13 million reads per sample).
Microbiome taxonomic profiling
Species-level profiling of the 815 samples was performed with both MetaPhlAn 3.0 (ref. 34) and MetaPhlAn 4.0 (ref. 46). Default parameters were used for both versions of MetaPhlAn, while specific databases to each version were used, mpa_v30_CHOCOPhlAn_201901 and mpa_vJan21_CHOCOPhlAnSGB_202103 for version 3 and 4, respectively. MetaPhlAn 3.0 taxonomic profiles were used to assess the presence and contribution of the previously identified 15 positively associated and 15 negatively associated species with dietary and cardiometabolic health markers2. MetaPhlAn 4.0 taxonomic profiles were analyzed to compare microbial compositions between participants and to determine alpha diversity indices, the number of detected species (observed richness). Microbiome taxonomic profiles were also analyzed to compare between-microbiome-sample dissimilarity (beta-diversity) using the Bray–Curtis dissimilarity measure.
Machine learning
We used the same machine learning framework developed by Asnicar et al.13 to assess the link of the microbiome compositions with the different dietary and metabolomic outcomes. Briefly, the machine learning framework is based on the random forest classification and regression algorithms and a 100-fold cross-validation approach with a 80/20 random splitting of the dataset. As training data, we used the differences in relative abundance between the 18-week and baseline time points of only microbial species. The classification task was evaluated using the area under the receiver operating characteristic curve, while the regression was evaluated by correlating the predicted values with the target values using the Spearman correlation coefficient.
Diet information
Participants completed the PREDICT FFQ online, at three separate time points throughout the study (0 weeks, 12 weeks and 18 weeks) to capture habitual dietary intake over the preceding month. The FFQ included 264 food and beverage items for which the participant selected frequency of consumption over the last month. Each survey item was accompanied by an USDA standard portion size, a textual description of the portion and a photograph of the item displayed on standard size tableware. The nutritional composition of each item was allocated according to the matching, or equivalent, item composition in the USDA database47; US nutrient intake, including macronutrient and micronutrient data, was calculated per participant. Submitted FFQs were excluded if more than ten food items were left unanswered, or if the total energy intake estimate derived from the FFQ as a ratio of the individual’s estimated basal metabolic rate (determined using the Schofield et al.’s equation48) was more than 2 s.d. outside the mean of this ratio (less than 0.15 or more than 2.04). Food energy density was calculated as the ratio between food energy (kcal) and food weight (g), excluding caloric (such as milk and juices) and noncaloric beverages28.
Safety
Adverse events were reported to the study coordinator, and were reviewed by the principal investigator and medical director. All adverse events were documented in line with IRB guidelines. The dietary intervention was anticipated to cause none to minimal discomfort. Some people may be affected by a small change in diet, for example, they may experience gas or bloating after eating the standardized test meals.
Sample size calculations
The study was powered on a sample size of 150 participants per group (n = 300) at 90% power and P −1 between-group difference in TG (endpoint change from baseline). An s.d. of 0.55 mmol l−1 was assumed on the basis of earlier data49. The same sample size was also powered to detect a 0.30 mmol l−1 change in LDL-C at 90% power and P −1 (ref. 49). Given two primary outcomes, statistical significance was defined by P
Statistical analysis
Analyses were carried out using v.4.0.2 of R and Python v.3.9.7. Pandas v.1.1.3, NumPy v.1.23.5 and SciPy v.1.11.1 were used to manage and preprocess data. Analyses of 18-week changes in primary and secondary outcomes were conducted based on an ITT (n = 347). We conducted a per-protocol analysis using the data collected from participants who returned to their endpoint visit as prespecified in the protocol (18 ± 2 weeks) (n = 225; 65% of the ITT cohort). An average of the two clinical blood chemistry baseline samples was used as the baseline measure for each participant. The primary outcome was the 18-week changes from baseline. The comparison between treatments in continuous variables over time was performed using repeated measures analysis ensuring that all ITT participants randomized with baseline information were included in the analysis and analyzed according to the original treatment assignment. The model evaluates the interaction between time (within-subject factor) and diet treatment (between-subject factor) with diet treatment, time, age and sex included as fixed effects along with a random effect for participants. The intervention effect was the coefficient for the interaction term in the model and the associated 95% CIs. The simple main effects of differences between the two diet groups were also assessed. For outcomes that were not normally distributed, outcomes were log10-transformed and tested for normality using the Shapiro–Wilk test. Given two primary outcomes, statistical significance was defined by P
We assessed gut microbiome composition using species-level taxonomic profiles of participants with longitudinal sampling available. The ITT cohort was restricted to 118 and 112 individuals for the control and PDP groups, respectively. For each individual, we calculated the within beta-diversity using the Bray–Curtis dissimilarity index between the longitudinal samples available. For the baselines (week −1 or week 0), when two samples were available for the same individual, we considered the one with the highest number of preprocessed reads. As reference beta-diversity variability for comparison with the week 12 and week 18 samples, we considered the values calculated in each individual with the two baseline samples available (both week −1 and week 0). Bray–Curtis dissimilarities of the longitudinal samples of the same individuals between control and PDP groups were tested using a paired, one-sided Wilcoxon rank-sum test, while across-intervention groups were tested using a Kolmogorov–Smirnov stochasticity parameter (KSp). As we previously identified microbial bacterial species associated with favorable and unfavorable cardiometabolic risk markers13, we tested differences between the two intervention groups. We tested statistically significant differences in terms of relative abundance values for favorable and unfavorable species between groups using a Mann–Whitney–Wilcoxon test (MWWp) and reported the magnitude and direction of change using a log2 fold change.
We performed a subgroup analysis based on dietary adherence to determine whether highly adherent participants differed across treatments. We identified adherent control participants (top 30% of participants based on the HEI score, a measure of adherence to USDA dietary guidelines) and compared them to adherent PDP participants (top 30% of participants based on a personalized diet quality score). Adherence to the ZOE program was classified based on a mean personalized diet score throughout the study duration. A minimum of 4 days of logged diet data meeting sex-specific caloric cutoffs (females, 500–5,000 kcal or more per day; males, 500–8,000 kcal or more per day) was required per month to ensure high quality and quantity logging. Low adherent participants were classified as the bottom 30th percentile of participants (mean personalized day score of 58 or lower); highly adherent participants were the top 30th percentile (mean personalized day scores of 67 or greater); moderately adherent participants fell in the middle (mean personalized day scores of 59–66). We also conducted a within-PDP analysis to investigate whether participants with good adherence (top 30%) to the PDP personalized dietary advice showed greater improvements in health outcomes compared to those with poor adherence (bottom 30%). Sex-based analysis was not performed because of small sample sizes. Excel v.16.82 and Microsoft Office were used for data and table formatting.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.