To trace the knowledge about food, chemical, and disease interactions, we have shown the creation of a KG centered around the impact of different foods and chemicals on CVDs, and the other targeting the composition of the selected food item “milk”, as well as its beneficial and detrimental effects on different NCDs. For this purpose, three NLP pipelines, called FooDis, FoodChem, and ChemDis, were combined to extract “food-disease”, “food-chemical”, and “chemical-disease” relations from textual data. Semantically, we distinguish two relations between food-disease and chemical-disease entity pairs, which are “treat” and “cause”. In the case of food-chemical entity pairs, we extracted only one relation which is “contains”. All three pipelines were executed twice, on two different corpora, one that was collected for CVDs and one collected for milk products. In both use cases, the searched keywords were selected by domain experts. In the CVDs case, a more general keyword was selected “heart disease food”, since we would like to retrieve broader aspects between different cardiovascular events and food products. This ends up with 9984 abstracts. In the milk use case, three keywords were selected by the domain experts i.e., “milk composition”, “milk disease”, and “milk health benefits”.
Table 1a presents the number of abstracts that were retrieved and used in the analysis for both use cases, together with the keywords used to retrieve them, while Table 1b presents the number of relations that were extracted for both use cases.
Table 1 Number of processed paper abstracts and number of extracted relations for each case study.
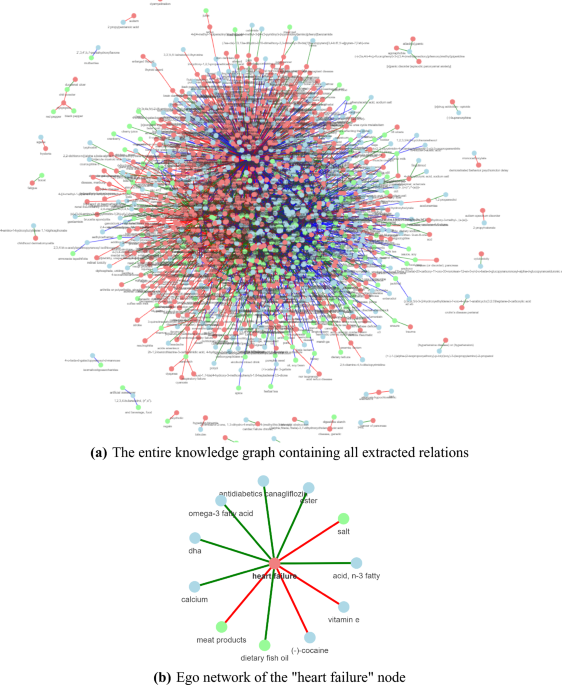
Figure 1a features the KG constructed by running the three pipelines for the two application use cases. The same nodes are grouped together by normalizing the extracted food, chemical, and disease entities.
Figure 1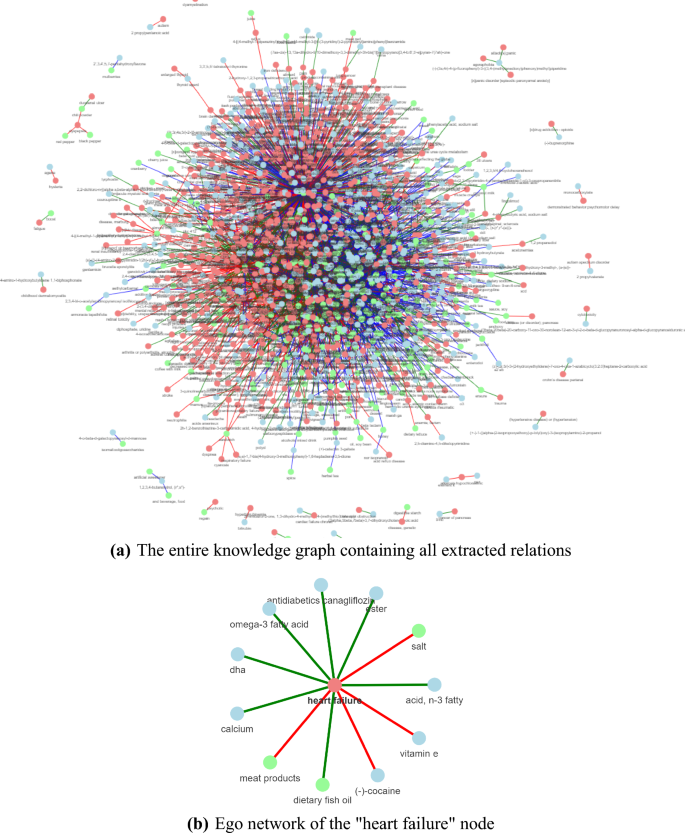
Knowledge graph constructed using the FooDis, FoodChem and ChemDis pipelines. The nodes in green represent the food entities, the nodes in blue represent the chemical entities, and the nodes in red represent the disease entities. The red, green, and blue edges represent the “cause”, “treat” and “contains” relations, respectively. The figures have been generated using the pyvis python library42, version 0.1.8.2.
To go into more detail how the KG is constructed, in Fig. 1b we present an example using the relations extracted for the “heart failure” disease entity. The green nodes, “meat products”, “salt” and “dietary fish oil” represent the food entities for which the FooDis pipeline extracted a relation with the “heart failure” disease entity, meaning that they have some effect on its development or treatment. In particular, the red edges connecting the “heart failure” disease entity and the food entities “meat products”, and “salt” indicate that the pipeline identified a “cause” relation, i.e. meat products, and salt can contribute to the occurrence of heart failure. On the other hand, the green edge between the “dietary fish oil” entity and the “heart failure” disease entity indicates a “treat” relation, i.e. the pipeline identified that dietary fish oil has a beneficial effect to heart failure. Similarly, the ChemDis pipeline identified that the chemical entities “DHA”, “ester”, “acid, n-3 fatty”, “antidiabetics canagliflozin”, “omega-3 fatty acid” and “calcium” can be used for treating “heart failure”, while the chemical entities “(-)-cocaine” and “vitamin E” can contribute to the development of “heart failure”. Table 2 presents the supporting sentences from scientific abstracts from which the relations were extracted and further used for constructing the graph presented in Fig. 1b. Next, such graphs are connected based on the same entities to link the information from different abstracts. Further, to validate the extracted information, domain experts were involved to check the extracted relations for both use cases.
Table 2 Supporting sentences for the relations of entity “heart failure” to different food and chemical entities.
Use case: cardiovascular diseases
For the CVDs use case, a highly-skilled domain expert (an MD with more than 40 years of working experience in cardiology) evaluated the extractions from the three pipelines. The relations that were evaluated are extracted after the “Final relation determination” step from the FooDis, FoodChem and ChemDis pipelines. All three pipelines utilized here follow the same workflow. Each extracted relation is determined by all sentences where information about it is presented. We called them “supporting sentences”. The sentences can be from the same or different abstracts, since information about the same relation can be investigated in different papers.
Domain expert evaluation
Each pipeline provides the result as a 6-tuple i.e., (name of the first entity, named of the second entity, synonyms for the first entity, synonyms for the second entity, relation, supporting sentences), which is further evaluated by the domain expert. The domain expert was asked to assign a binary indicator of the truthfulness of the relation. The pipelines were then evaluated by taking the mean of the correctness indicators assigned by the annotator for each relation and pipeline, which we refer to as the precision in the remainder of this section. In particular, if a pipeline extracted three relations, and the expert marked two of these as correct (binary indicators 1,0,1), the reported precision would be 0.66.
Figure 2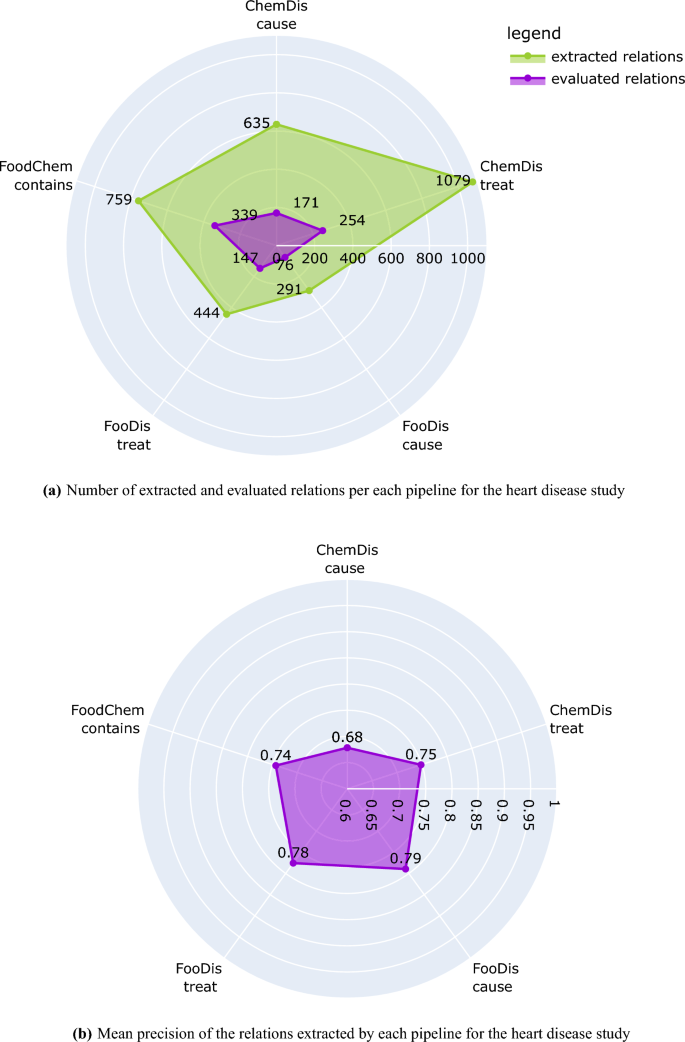
Number of extracted and evaluated relations and mean precision of each pipeline for the heart disease study. The plots have been generated using the plotly python library43, version 5.7.0.
Figure 2a presents the number of relations extracted by each of the pipelines for the CVDs study, and the number of relations that the domain expert evaluated. We need to point out here that all extracted relations were provided to the domain expert, however, the evaluation has been performed only on those relations for which the domain expert has expert knowledge. Because of this, the human evaluation process covers 44% of the “contains” relations extracted by the FoodChem pipeline, 33% of the “treat” relations extracted by the FooDis pipeline, 26% of the “cause” relations extracted by the FooDis pipeline, 26% of the “cause” relations extracted by the ChemDis pipeline, and 23% of the “treat” relations extracted by the ChemDis pipeline.
The mean precision of each of the pipelines (FooDis, ChemDis, and FoodChem) in the CVDs use case is presented in Fig. 2b. From it, the FooDis pipeline achieves the highest precision of 0.79 for the “cause” and 0.78 for the “treat” relation. The lowest precision of 0.68 is achieved by the ChemDis pipeline for the extraction of the “cause” relation.
Since the three pipelines extract a relation based on supporting sentences, in the Supplementary Materials, we have presented the distribution of the number of relations versus their number of supporting sentences.
All of the pipelines extract more than 74% of the relations based on a single supporting sentence. The ChemDis and FoodChem pipelines can find a larger number of supporting sentences for some relations compared to the FooDis pipeline. In particular, the ChemDis pipeline can find up to five supporting sentences to identify “cause” relations and up to 14 supporting sentences to identify “treat” relations, while the FooDis pipeline uses up to three, and four supporting sentences for the “cause” and “treat” relations, respectively.
Next, to see how the mean precision is affected by the number of supporting sentences, we analyze for each semantic relation separately. The results are presented in Supplementary Materials. From the conducted analysis, we can conclude that the mean precision is proportional to the number of supporting sentences. Almost for all relations, a precision of 1.00 is reached when the number of supporting relations is sufficiently high. This indicates that when the number of supporting sentences for a relation increases, there is an agreement between the domain expert validation and the result provided by our pipelines, with some exceptions listed in the Supplementary Materials.
Error analysis
Next, we analyze the types of false discoveries produced by FooDis, FoodChem, and ChemDis pipelines.
Figure 3 features the relations with the highest number of supporting sentences for four chemical entities: “carbohydrates”, “fatty acid”, “sodium” and “vitamin d”. Here the results for the selected chemical entity from the two pipelines that deal with chemical entities (i.e., ChemDis and FoodChem) are presented. The green bars refer to the number of sentences in which the relation was correctly identified, while the purple plots refer to the number of false positive sentences for that relation, i.e. sentences where the relation was identified, however, it was marked as incorrect by the experts.
For the “carbohydrates” entity, the ChemDis pipeline produced the false positive relation “carbohydrates-treat-cardiomyopathy” when the supporting sentences suggested that a low-carbohydrate diet is recommended for treating cardiomyopathy. In this case, the pipeline fails to identify that a reduction of the chemical entity is required to treat the disease. In addition, the FoodChem pipeline produces a false discovered relation “bulk-contains-carbohydrates”, when the supporting sentence was saying that these two entities are contained in another entity, “dry beans”. For the “fatty acid” chemical entity, the ChemDis pipeline produced the false positive relation “fatty acid-cause-dysfunction endothelial”, when the supporting sentence was saying that increased fatty acid levels and endothelial dysfunction were contributing to the development of another disease, “sepsis”. The FoodChem pipeline produced the false entities, “wine-contains-fatty acid” and “acid fatty trans-contains-fatty acid”. In the first case, the two entities were co-occurring in the supporting sentence without any relation, while in the second one, the sentence was saying that trans fatty acids are a subcategory of fatty acids. In the case of the “sodium” chemical entity, most of the sentences extracted by the ChemDis pipeline express the correct relation, however, sodium is incorrectly extracted as a partial match of the entity “Sodium-glucose co-transporter 2 inhibitors (SGLT2is)”. In the case of “vitamin d”, all of the false positive “cause” relations extracted by the ChemDis pipeline are due to the pipeline not recognizing that the deficiency of the vitamin was causing the diseases.
Figure 3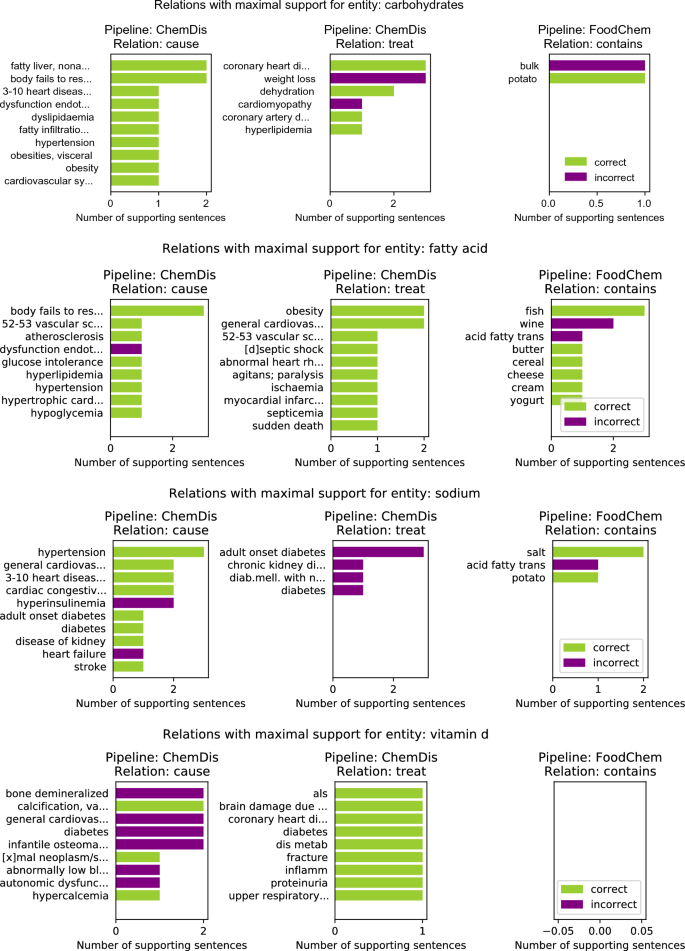
Top 10 “cause”, “treat”, and “contains” relations with maximum number of supporting sentences for four chemical entities: “carbohydrates”, “fatty acid”, “sodium” and “vitamin d”. The entities in the rows of the ChemDis pipeline are diseases caused or treated by the chemical, while the entities in the rows of the FoodChem pipeline are food entities in which the chemical is contained.
Figure 4 features the top 10 relations with a maximal number of supporting sentences for three disease entities. Here, we present the results from pipelines that are dealing with disease entities (i.e., FooDis and ChemDis). For the “general cardiovascular disorders” entity, the pipelines extracted the relations “dietary vegetable-cause-general cardiovascular disorders”, “acid, saturated fatty-treat-general cardiovascular disorders”, “acid fatty polyunsaturated-cause-general cardiovascular disorders”, “cholesterol-treat-general cardiovascular disorders” due to the fact that the pipelines were not able to recognize that the sentences were referring to the reduction of these food or chemical entities affecting the disease development or treatment of the general cardiovascular disorders. This is also the reason for false positive relations extraction for the other two disease entities featured in the figure.
Figure 4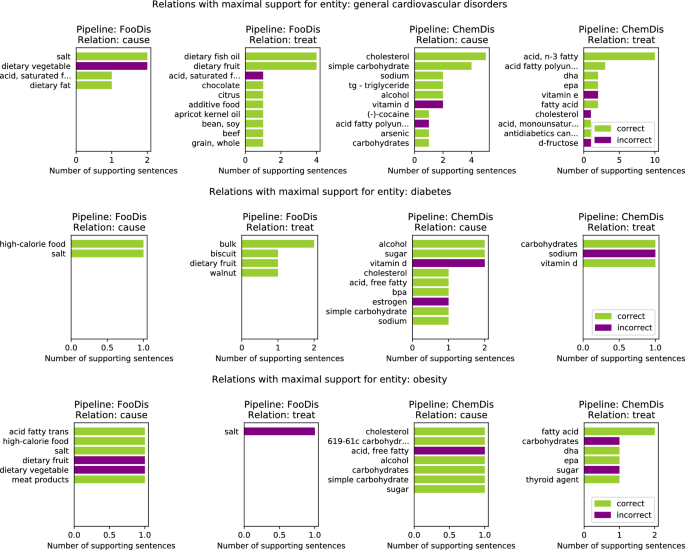
Top 10 “cause” and “treat” relations with maximal number of supporting sentences related to three disease entities: “general cardiovascular disorders”, “diabetes”, and “obesity”. The entities listed in the rows of the FooDis pipeline are food entities, while the entities listed in the rows of the ChemDis pipeline are chemical entities, that cause or treat the specified disease.
Use case: milk
For the use case related to the composition and health effects of milk, two highly-skilled domain experts evaluated the results from all three pipelines: a chemist and a food and nutritional scientist.
Domain expert evaluation
From the 33,111 processed abstracts related to the milk case study, the three pipelines extracted a total of 6792 relations, from which 5139 were evaluated by the two domain experts. We need to point out again that all extracted relations were provided to the domain experts, however, they evaluated only those relations for which they have domain expertise. Figure 5a features the number of relations extracted by each pipeline for the milk case study, and the number of relations the experts evaluated. The highest number of evaluated relations were the “contains” relations extracted by the FoodChem pipeline, and the experts were able to evaluate 96% of them (2849 out of 2754). The experts also evaluated 73% of the “treat” and 78% of the “cause” relations produced by the FooDis, 34% of the “cause” relations, and 35% of the “treat” relations produced by the ChemDis pipeline.
Figure 5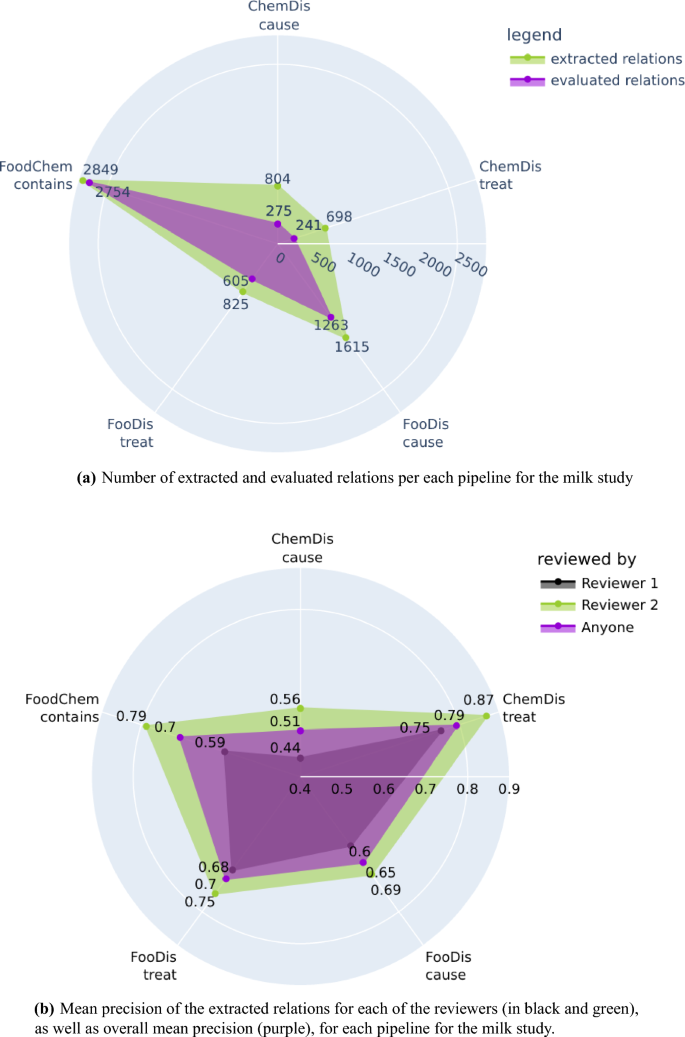
Number of extracted and evaluated relations and mean precision of each pipeline for the milk study. The plots have been generated using the plotly python library43, version 5.7.0.
The mean precision for each of the five semantic relations for both domain experts is presented in Fig. 5b separately. In addition, we have also presented the mean precision for each type of relation by averaging the precision across both domain experts. From the figure, we can see that the first domain expert, who evaluated the relations which were supported by a single sentence, identified more incorrect relations than the second domain expert, who evaluated the relations supported by multiple sentences.
The overall mean precision for each of the five relations averaged across both domain experts are as follows:
0.51 for the “cause” relation extracted by the ChemDis pipeline,
0.79 for the “treat” relation extracted by the ChemDis pipeline,
0.65 for the “cause” relation extracted by the FooDis pipeline,
0.70 for the “treat” relation extracted by the FooDis pipeline,
0.70 for the “contains” relation extracted by the FoodChem pipeline.
The error analysis for the milk case study followed the same procedure as for the heart disease study and resulted in similar findings, presented in the Supplementary Materials.