Mice
All mice, including wild-type C57BL/6J, Lepr-Cre (stock no. 008320) and Penk-Cre (stock no. 025112) mice, were obtained from the Jackson Laboratory. To visualize Lepr-positive neurons, Lepr-Cre mice were crossed with Ai14 mice (stock no. 007908; tdTom reporter line; Jackson Laboratory). All transgenic mice for behavioral experiments were backcrossed to wild-type C57BL/6J mice for multiple generations. A similar number of both male and female mice (10–13 weeks old) were used for body weight and food intake measurements, behavioral experiments, immunohistochemistry, RT–qPCR, FISH and anatomical tracing. Adult male and female mice (10–13 weeks old) were used for ex vivo slice electrophysiology, metabolic cage analysis and glucose tolerance tests. We initially examine the behaviors of males and females separately, but we didn’t find any significant difference. Mice were housed on a 12-h light/dark cycle with standard bedding in a temperature-controlled and humidity-controlled room (~21 °C and 42% humidity). All behavioral procedures were performed during the light cycle.
Experimenters were blind to the group allocation and outcome assessment. For data analysis, primary experimenters were not blind due because the experimental conditions (for example, stress paradigm, food exposure) were obvious to the researchers, but the analysis was carried out without subjective bias. All experiments were approved by the Institutional Animal Care and Use Committee of the Virginia Polytechnic Institute and State University and the University of California, San Diego.
Early-life trauma procedures
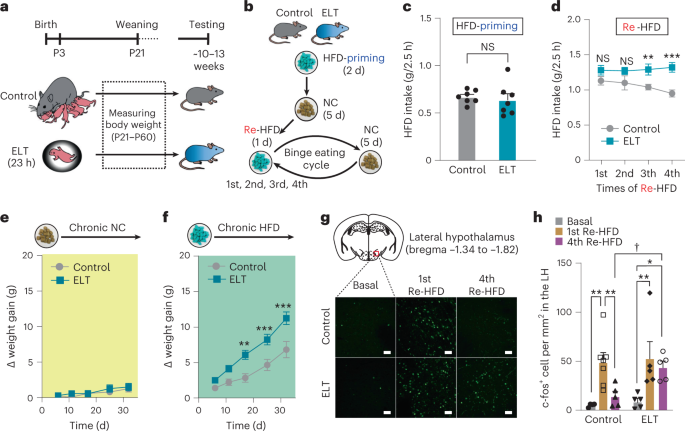
The ELT paradigm was adopted from previously published methods with minor modifications59,60. Briefly, pregnant female mice were individually housed when they were 14–16 d pregnant. From P3 to P4, ELT pups were separated from both their dam and littermates for 23 h. During this separation, ELT pups were placed individually in a divided small chamber with clean bedding and transferred to an incubator, which was maintained at 32 ± 1 °C. At the end of the separation period, ELT pups were reunited with the dam and littermates. Control pups remained undisturbed in the maternal nest. All pups were weaned at P21 and housed in groups of three to five of the same gender until the start of the experiments.
Binge-like eating paradigm
The binge-like eating behavior in mice was monitored using the repetitive intermittent-access schedule of a HFD (60% kcal% fat; D12492, Research Diets) with minor modifications of the previous publication30. The mice were fed standard NC (18% kcal% fat; no. 2918, Harlan-Teklad) until training and assessment of binge-like eating. Mice (10–13 weeks old) were singly housed and subjected to an initial 2 d of free access to HFD, then only NC was available ad libitum for the following 5 d. On the next day, mice were exposed to a HFD again (Re-HFD) and the food intake level was monitored 2.5 h later. After 24 h, the HFD was removed and replaced with NC only, thus completing the 1st binge eating cycle of Re-HFD. For repetitive cycles, mice were subjected to a 6-d NC-only period, which was followed by Re-HFD for 24 h. Foods were always available ad libitum with no food restrictions.
Body weight and food intake measurements
Body weight changes or food intake of mice (10–13 weeks old) were recorded in their home cages for 3–4 weeks or 5 d of access to either NC or HFD. Food intake was assessed by subtracting the amount of food remaining in cages from the weight of foods initially provided.
Metabolic phenotyping experiments
Mice were fed with a HFD and subjected to metabolic cage analysis to evaluate energy expenditure. Metabolic parameters including O2 consumption, CO2 production and respiratory exchange ratio were recorded by Comprehensive Lab Animal Monitoring System (Columbus Instruments)61 for three consecutive days and nights, with at least 24 h of an adaptation period before data recording.
Serum leptin and corticosterone measurement
Mice were anesthetized with isoflurane and decapitated. Whole trunk blood samples were collected in 1.5 ml microcentrifuge tubes and allowed to clot at room temperature for 30 min. Tubes were then spun for 15 min at 3,000g at 4 °C. Supernatant containing clear serum was stored at −80 °C until enzyme-linked immunosorbent assay to measure serum leptin and corticosterone using an assay kit (EMD Millipore EZML-82K, and Cayman 501320, respectively).
Glucose tolerance test
Glucose tolerance tests were done after mice underwent a fasting period of 16 h with water ad libitum. Mice were subsequently given i.p. injections of 100 mg ml−1 d-glucose (2 g per kg body weight). Five microliters of blood collected from the tail vein was dropped onto a glucose test strip (Breeze 2 blood glucose meter kit, Bayer). Blood glucose was determined at 0, 40 and 120 min after the injection.
Locomotion
Locomotion was assessed in an open field arena (44 × 44 × 44 cm3). Mice were placed individually in the arena and allowed to explore freely for 15 min. The activity was monitored using a webcam mounted above the arena and analyzed by tracking software (Viewer 3.0, BIOBSERVE).
Open field test
Each mouse was placed in the open field arena (44 × 44 × 44 cm3) and allowed to move freely for 15 min. The open field area was subdivided into two zones, a center (20 × 20 cm2) and a periphery. For the chemogenetic experiment, CNO (2 mg per kg body weight, i.p.) was administered 30 min before the open field test session. The movement of mice was monitored with a webcam and analyzed by tracking software (Viewer 3.0, BIOBSERVE).
Elevated plus maze
The elevated plus maze consisted of two open arms, two closed arms and a center elevated to a height of 30.5 cm above the ground. Mice were placed in the center and allowed to explore the space for 5 min. For the chemogenetic experiment, CNO (2 mg per kg body weight, i.p.) was administered 30 min before the elevated plus maze session. The movement of mice was analyzed by tracking software (Viewer 3.0, BIOBSERVE).
Novel object recognition test
Mice were habituated to the open field arena (44 × 44 × 44 cm3) in the absence of objects for 30 min a day before the training session. During the training session, two identical objects were placed in each corner of the arena, and mice were allowed to explore for 10 min. Twenty-four hours after training, mice were placed in the arena where one of the two objects was replaced with a novel object having a different color and shape. All movements of mice were monitored with a webcam for 10 min, and the recognition time in each object area (2 cm around the object) was measured by tracking software (Any-maze). Discrimination rate was calculated as ((time spent in a novel object area) / (time spent in a novel object area + time spent in a familiar object area) × 100(%))42,62.
Rotarod test
For the fixed speed rotarod test (17 r.p.m.), each mouse was given two practice trials and then placed on the rotating cylinder in a rotarod apparatus (Ugo Basile). The latency to fall off the rotarod was recorded with a 3-min cutoff per session63.
Immunohistochemistry
For c-fos immunoreactivity, mice were individually housed before the training and assessment of binge-like eating behavior. After the HFD withdrawal period (5 d), a HFD was reintroduced to mice for 2.5 h. Subsequently, the mice were then anesthetized with isoflurane and transcardially perfused with 4% paraformaldehyde (PFA). Brains were extracted and post-fixed overnight in 4% PFA. Coronal sections (50 μm) were washed in PBS containing 0.3% Triton X-100 (PBS-T, pH 7.4) and incubated in 1% BSA (vol/vol) in PBS-T for 1 h. The sections were immunostained using a rabbit anti-c-fos antibody (1:5,000 dilution; Cell Signaling Technology) applied overnight in PBS-T, at room temperature. On the next day, sections were washed in PBS-T and incubated in a 1:500 dilution of Alexa Fluor Plus 488 anti-rabbit secondary antibody (Thermo Fisher Scientific) in PBS-T for 1 h. For Mch and Hcrt immunostaining, rabbit anti-Mch (1:1,000 dilution; Phoenix Pharmaceuticals) and anti-Hcrt (1:1,000 dilution; Phoenix Pharmaceuticals) were applied. For phospho-STAT3 (pSTAT3) immunofluorescence staining, rabbit anti-pSTAT3 antibodies (1:500 dilution; Cell Signaling) were applied overnight at room temperature. Thereafter, the sections were rinsed with PBS-T and incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:1,000 dilution; Cell signaling Technology) in PBS-T. Subsequently, sections were washed and incubated for 10 min with a 1:50 dilution of tyramide signal amplification (TSA) Plus Fluorescein (TSA Plus kit, PerkinElmer Life Sciences) to enhance the pSTAT3 signal. Sections were rinsed with PBS-T and mounted using a mounting medium. Images were acquired using a confocal microscope (Olympus FluoView FV1200) and quantitatively analyzed with ImageJ. For c-fos and pSTAT3 quantification, the neurons located lateral and up to 0.1 mm medial to the fornix or up to 0.2 mm above or below the fornix were considered to be in the LH. Cell counts were made within this reference area by applying equal thresholds to all images and using the ‘analyze particles’ function in ImageJ. For c-fos quantification in Lepr-Cre × Ai14 mice, c-fos and tdTom double-positive cells were counted. The percentage of double-positive cells was calculated among the total numbers of tdTom-expressing cells.
RT–qPCR
Mice were anesthetized with isoflurane and 250-µm slices were prepared in PBS, using a vibratome (Leica VT1200). The LH was microdissected bilaterally, and samples were immediately frozen on dry ice and stored at −80 °C before RNA isolation. Total RNA was extracted from dissected samples using a Hybrid-R RNA purification kit (GeneAll Biotechnology). Purified RNA samples were reverse transcribed by using the SuperScript-IV First-strand cDNA synthesis kit (Invitrogen). qPCR was performed by using TaqMan Gene Expression Assay Kit (Applied Biosystems). All TaqMan probes were purchased from Applied Biosystems and are as follows: Lepr (Mm00440181_m1), Hcrtr1 (Mm01185776_m1), Mchr1 (Mm00653044_m1), Hcrt (Mm01964030_s1), Pmch (Mm01242886_g1), Galr1 (Mm00433515_m1), Nts (Mm00481140_m1), Gal (Mm00439056_m1), Cartpt (Mm04210469_m1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Mm99999915_g1). Target amplification was performed by using ViiA 7 Real-Time PCR System (Applied Biosystems) with QuantStudio Real-Time PCR software v1.3. The relative mRNA expression levels were calculated via a comparative threshold cycle (Ct) method using GAPDH as an internal control: ΔCt = Ct (gene of interest) − Ct (GAPDH). The gene expression fold change, normalized to the GAPDH and relative to the control sample, was calculated as the 2−ΔΔCt methods64.
RNA in situ hybridization
Brains were rapidly extracted and flash frozen with isopentane (Sigma-Aldrich) chilled with dry ice in 70% ethanol. Coronal brain slices (16 µm) containing the LH were sectioned using a cryostat (Leica CM3050S) at −20 °C. Brain slices were mounted directly onto slides and stored at −80 °C until RNA in situ hybridization which was conducted using RNAscope probes (Advanced Cell Diagnostics, ACD). Slides were fixed in 4% PFA for 15 min at 4 °C and subsequently dehydrated for 5 min with 50%, 70% and 100% ethanol at room temperature. Sections were then incubated with a Protease IV solution for 30 min, and washed with PBS, before being incubated with probes for 2 h at 40 °C in the HybEZ oven (ACD). All probes used were commercially available: Mm-Lepr (402731), Mm-Hcrt (490461), Mm-Pmch (478721), Cre (312281), Mm-Cartpt (432001), Mm-Nts (420441) and Mm-Slc32a1 (319198). After washing with wash buffer, the signal was amplified by incubating tissue sections in amplification buffers at 40 °C. After the final rinse, DAPI solution was applied to the sections. Slides were visualized with a confocal microscope (Olympus FluoView FV1200).
Circuit mapping quantification
For LHLepr neuronal downstream connectivity quantitation, eGFP-labeled fibers and synaptic puncta were quantified as previously described with slight alterations65,66. Briefly, 50-µm coronal sections were obtained across the anteroposterior axis of the brain. Brain regions were determined by anatomical landmarks and based on the Paxinos and Franklin mouse brain atlas67. Images were taken with an Olympus FluoView FV1200 confocal microscope. Analysis was conducted in Fiji (ImageJ) and quantified as a percentage of the area of thresholded pixels normalized to either the HSV-DIO-Flp injection sites (Fig. 4c,f) or the vlPAG (Extended Data Fig. 7n,o). Qualitatively determined threshold values were obtained by determining the level that best mirrored the original image without introducing a background and maintained the consistency throughout animals.
Virus and shRNA generation
AAV was produced by transfection of 293 cells with three plasmids: an AAV vector expressing target constructs (EmGFP, EmGFP-Lepr shRNA, DIO-EmGFP, DIO-EmGFP-Lepr shRNA, saCas9, Lepr sgRNA (a gift from D. Kong), DIO-GCaMP6f, DIO-eGFP, DIO-Synaptophysin-eGFP, DIO-eNpHR3.0-eYFP, DIO-eYFP, fDIO-mCherry, fDIO-hM3D-mCherry, fDIO-hM4D-mCherry, fDIO-eGFP, fDIO-Kir2.1-eGFP, DIO-mCherry, DIO-mCherry-hM3D, DIO-mCherry-hM4D and DIO-mRuby2-TVA-RVG), AAV helper plasmid (pHELPER; Agilent) and AAV rep-cap helper plasmid (pRC-DJ, a gift from M. Kay). At 72 h after transfection, the cells were collected and lysed. Viral particles were then purified by an iodixanol step-gradient ultracentrifugation method. The iodixanol was diluted and the AAV was concentrated using a 100-kDa-molecular-mass-cutoff ultrafiltration device. The genomic titer was determined by qPCR. The AAV vectors were diluted in PBS to a working concentration of approximately 1012 viral particles per ml. To generate EnvA-pseudotyped glycoprotein (G)-deleted rabies virus expressing eGFP (RVΔG-eGFP), we followed a published protocol68. Plasmids expressing the rabies viral components, B7GG, BHK-EnvA and HEK-TVA cells were provided courtesy of E. M. Callaway. HSV-DIO-Flp was purchased from the Gene Delivery Technology Core at the Massachusetts General Hospital.
To construct shRNA against Lepr (NM_146146.3), oligonucleotides that contained 21-base-pair sense and antisense sequences (5′-AACTGATGAAGAGCAAGGGTT-3′) targeting Lepr were connected with a hairpin loop followed by a poly(T) termination signal. This shRNA oligonucleotide was ligated into BLOCK-iT POLII miR RNA-mediated interference expression vectors (Invitrogen) and then transferred to an AAV vector together with EmGFP. To test the efficacy of the shRNA, we stereotaxically injected AAVs expressing Lepr shRNA into the LH. Two weeks after injection, the virus-infected area labeled by EmGFP expression was dissected. Lepr mRNA levels were measured by qPCR and found to be reduced over 70% (Fig. 2d and Extended Data Fig. 3l).
Stereotaxic surgeries and histology
Lepr-Cre or wild-type mice (8–10 weeks old) were anesthetized with a mixture of ketamine (100 mg per kg body weight) and dexmedetomidine (0.5 mg per kg body weight). The mouse was mounted in a stereotaxic frame (David Kopf Instruments). Body temperature was kept stable by using a heating pad while recovering from anesthesia. Viral injections were targeted using coordinates based on the Paxinos and Franklin mouse brain atlas67.
For shRNA-related testing in behavior or electrophysiology experiments, viral preparations (AAV-EmGFP, AAV-EmGFP-Lepr shRNA, AAV-DIO-EmGFP, AAV-DIO-EmGFP-Lepr shRNA) in a 300–350-nl volume were injected bilaterally into the LH (bregma, anteroposterior −1.65 mm; lateral ±1.12 mm; dorsoventral −5.25 mm) at a slow rate (100 nl min−1) using a syringe pump. Mice were allowed 2 weeks to recover from the virus injections before starting of either behavioral tests or electrophysiological recording. Injection sites were confirmed in all animals by preparing coronal sections containing the desired plane, and animals with incorrect injection placement were excluded from analyses.
For the CRISPR–SaCas9 viral-based system, both the AAV carrying an sgRNA targeting the mouse Lepr locus (AAV-sgLepr38, gift from D. Kong) and the AAV expressing S. aureus Cas9 (SaCas9; AAV-hSyn-SaCas9-U6-sgRNA) were bilaterally co-injected into the LH of wild-type mice. Three weeks were given for viral expression before behavioral tests.
For in vivo Ca2+ imaging experiments, Lepr-Cre animals received a unilateral injection of AAV-DIO-GCaMP6f (250 nl) into the LH. After 10–15 min of viral infusion, a sterile 20-gauge needle was slowly lowered into the same site to a depth of −4.65 mm from the cortical surface to clear a path for the implantation of a GRIN lens. The snap-in imaging cannula (model L-V; 500 µm in diameter; 5.66 mm in length; Doric Lenses) with GRIN lens was then implanted above the virus injection site. The target depth of the lens was adjusted to 100 µm above the viral injection site. The implanted imaging cannula and focusing ring were secured to the skull with an initial layer of adhesive cement (C&B Metabond; Parkell) followed by a second layer of dental cement (Ortho-Jet; Lang). In vivo Ca2+ imaging tests were performed 6 weeks after the implantation of the image cannula.
For anatomical output mapping, either AAV-DIO-eGFP or AAV-DIO-synaptophysin-eGFP (300 nl) were unilaterally injected into the LH of Lepr-Cre mice. Mice were euthanized 2 weeks after the viral injection for examining the outputs of LHLepr neurons. For input mapping experiments with EnvA-pseudotyped rabies virus, we first unilaterally injected 300 nl of AAV-DIO-mRuby2-TVA-RVG into the vlPAG (bregma, anteroposterior −4.62 mm; lateral ±0.3 mm; dorsoventral −2.82 mm) of Penk-Cre mice. Two weeks later, mice were again anesthetized as previously described and injected with EnvA-pseudotyped RVΔG-eGFP into the same site, vlPAG. The mice were euthanized 5 d after the last injection for input mapping analysis.
For optogenetic behavioral experiments, Lepr-Cre animals were bilaterally injected with 350 nl of AAV-DIO-eYFP or AAV-DIO-eNpHR3.0-eYFP into the LH. Bilateral chronic optic fibers (200 µm in diameter, 0.22 NA; Doric Lenses) were implanted above the downstream targets of LHLepr neurons in either vlPAG or VTA (bregma, anteroposterior −3.15 mm; lateral ±0.4 mm; dorsoventral −4.42 mm). Implanted fibers were adhered to the skull with adhesive and dental cement as described above. Lastly, sutures or sterile tissue adhesive (Vetbond; 3M) was used to close the incision. Three weeks were given for viral expression in terminals before behavioral tests. Upon completion of the behavioral experiment, viral injections and fiber placement were confirmed.
For the manipulation of LHLepr neurons in a projection-specific manner, Lepr-Cre animals were bilaterally injected with 350 nl of AAV-fDIO-mCherry, AAV-fDIO-mCherry-hM3D, AAV-fDIO-mCherry-hM4D, AAV-fDIO-eGFP or AAV-fDIO-Kir2.1-eGFP into the LH. During the same surgery session, HSV-DIO-Flp (350 nl) was injected into either vlPAG or MPA (bregma, anteroposterior −0.11 mm; lateral ±0.25 mm; dorsoventral −5.35 mm). For manipulation of vlPAGPenk neurons, AAV-DIO-mCherry, AAV-DIO-mCherry-hM3D, AAV-DIO-mCherry-hM4D, AAV-DIO-eGFP or AAV-DIO-Kir2.1 was bilaterally injected (300 nl) into the vlPAG of Penk-Cre mice. Three weeks were given before behavioral testing.
For microinjection of the leptin or PESLAN into the LH, a guide cannula (26-gauge, 6-mm long; Plastics One) was chronically implanted in this brain region (bregma, anteroposterior −1.65 mm; lateral ±1.12 mm; dorsoventral −5.05 mm). Implanted cannulae were secured to the skull as described above, and then obturators were placed in the guide cannulae. Behavioral testing was performed 2 weeks after the implantation. Upon completion of all behavioral experiments, viral injections or fiber/cannula placements were confirmed. Mice with off-target expression and/or off-target implant tip location were excluded from the final analyses.
After each experiment, the extent of viral transduction spread was examined at the conclusion and viral transduction was validated whether it was limited to the LH without off-target effects in other adjacent areas such as the Arc, VMH and DMH. The inclusion criteria were applied when the virally labeled neurons were located in the LH reference area; that is, lateral and up to 0.1 mm medial to the fornix or up to 0.2 mm above or below the fornix. If the viral expression was found outside this reference area or the viral transduction was weak in the LH (covering less than 50% of the total LH area), the mice were excluded from the final dataset, which was determined by two experimenters who were blinded to the experimental design.
Optogenetic stimulation
A 3-m-long fiber-optic patch cord (Doric Lenses) was connected to the chronically implanted optic fiber and suspended above the behavioral testing area to allow animals to move freely while receiving laser illumination. For eNpHR3.0-mediated inhibition, the patch cord was connected to a 593-nm laser (OEM Laser Systems). Bilateral inhibition of the LHLepr → vlPAG or LHLepr → VTA circuit was achieved by delivering continuous yellow light for 30 min. The power of the light at the tip of each optic fiber was adjusted to 10 mW. On the day of testing, each mouse was then placed in an experimental chamber (22 × 22 × 22 cm3) with a transparent acrylic bottom, which is located 40 cm above the webcam. After a 30 min habituation to the chamber and the patch cord, either HFD or Re-HFD was introduced to the mouse with optical stimulation (ON) for 30 min. All behaviors were recorded with a bottom view69 to analyze the latency to the first eating bout, eating duration, and physical contact to the food.
In vivo Ca2+ imaging in freely moving mice and data analysis
For in vivo Ca2+ imaging experiments, mice were exposed to HFD priming or Re-HFD. On the testing day, mice were habituated to a clear glass chamber (25 cm in height, 20 cm in diameter) for 30 min with the head-mounted microscope body attached to the top of the imaging cannula. To monitor the GCaMP6f fluorescence change during binge-like eating behaviors, Re-HFD was presented for 2 min with a previous recording of baseline fluorescence for the same period of time. Mice eating behaviors were recorded by a web camera concurrently. Images were acquired at 8 frames per second with an average exposure time of 125 ms using Doric Neuroscience Studio software (version 5.3.1.2; Doric Lenses). LED power was maintained at 30% with analog gain 2. All image analysis with ΔF/F0 was performed with a Doric Neuroscience Studio software (Doric Lenses). To remove movement artifacts, individual image frames were aligned using a single frame as a reference, and then background fluorescence was removed from the aligned images. Regions of interest corresponding to cell bodies were determined using an automated cell-finding function and were visually inspected to ensure accuracy. Neural Ca2+ dynamics (ΔF/F0) were presented as a heat map using customized MATLAB code. The ΔF/F0 values were smoothed with a Gaussian filter and sorted in descending order after the introduction of Re-HFD. For the classification of neurons, the basal level of ΔF/F0 was determined for 2 min before the food presentation. Cells were considered as up or down if the ΔF/F0 value during the first 2 min after presenting Re-HFD was higher than basal ΔF/F0 + standard deviation (σ) or lower than basal ΔF/F0 − σ, respectively. Cells with average fluorescence between basal ΔF/F0 + σ and ΔF/F0 − σ were categorized as non-response. To detect individual Ca2+ transients, we first processed the ΔF/F0 time series to remove slow drifts, estimated by a median filter of 10 s in width. Next, the median absolute deviation (MAD) of the entire time series was computed. Ca2+ transients were extracted by sequentially detecting each upward transient that exceeded a 6-MAD threshold (equivalent to 4 σ assuming normal distribution) and following the previous event by an interval no shorter than 2 s. For correlating LHLepr neuronal activity with eating bouts, Ca2+ transients of cells from individual animals aligned to the first eating bout onset. For 1.5 min after the first eating bout, the number of cells producing at least one Ca2+ transient that occurred at the onset of eating bouts in the presence of HFD were counted. The percentage of the correlated or non-correlated cells was calculated among the total numbers of LHLepr neurons that were detected during the image session. It was challenging to get a large number of the LHLepr neurons per animal in this area for clustering analysis. In addition, because of the difficulty of tracking same neurons over a long period of time, our analysis focused on the changes within sessions.
For in vivo Ca2+ imaging experiments with either a novel object (for example, Lego brick) or a social stimulus (for example, mice urine from the opposite sex conspecifics), the individual mouse was habituated to a clear glass chamber (25 cm in height, 20 cm in diameter) for 30 min with the head-mounted microscope body attached to the top of the imaging cannula. The basal level of average cell fluorescence (ΔF/F0) was determined for 2 min before either novel object or mice urine application.
Ex vivo electrophysiology
Coronal brain slices (300 µm) containing the LH were prepared using a vibratome (Leica VT1200S), in a solution containing: 110 mM choline chloride, 25 mM NaHCO3, 1.25 mM NaH2PO4, 2.5 mM KCl, 7 mM MgCl2, 25 mM glucose, 0.5 mM CaCl2, 11.6 mM ascorbic acid and 3.1 mM pyruvic acid, saturated with 95% O2/5% CO2. Slices were then allowed to recover at 30 °C for 20 min, and subsequently at room temperature, in a solution containing: 118 mM NaCl, 26 mM NaHCO3, 11 mM glucose, 15 mM HEPES, 2.5 mM KCl, 1.25 mM NaH2PO4, 2 mM pyruvic acid, 0.4 mM ascorbic acid, 2 mM CaCl2 and 1 mM MgCl2, saturated with 95% O2/5% CO2. Slices were maintained at room temperature for at least 1 h until transferred to a recording chamber perfused with: 119 mM NaCl, 26.2 mM NaHCO3, 11 mM glucose, 2.5 mM KCl, 1 mM NaH2PO4, 2.5mM CaCl2 and 1.3 mM MgCl2, saturated with 95% O2/5% CO2, and delivered at 2 ml min−1 at 30 ± 1 °C. For all recordings, patch pipettes (3–5 MΩ) were pulled from borosilicate glass (G150TF-4; Warner Instruments) with a DMZ Universal Electrode Puller (Zeitz Instruments) and filled with appropriate intracellular solutions. Neurons were visualized with differential interference contrast optics or epifluorescence (Olympus). Recordings were made with a MultiClamp 700B amplifier and pClamp10 software (Molecular Devices). Data were low-pass filtered at 1 kHz and digitized at 10 kHz with a digitizer (Digidata 1440; Molecular Devices). Series resistance was monitored and cells that displayed a change > 20% throughout recording were excluded.
To measure the E/I ratio using voltage-clamp recording, pipettes were filled with a Cs-based intracellular solution, containing: 115 mM Cs+-methanesulphonate, 10 mM HEPES, 1 mM EGTA, 1.5 mM MgCl2, 4 mM Mg2+-ATP, 0.3 mM Na+-GTP, 10 mM Na2-phosphocreatine, 2 mM QX 314-Cl and 10 mM BAPTA-tetracesium (295 mOsm, pH 7.35). Electrically evoked excitatory postsynaptic and inhibitory postsynaptic currents (eEPSCs and eIPSCs, respectively) were recorded from identified LHLepr neurons at −70 mV (for EPSCs) and 0 mV (for IPSCs). The E/I ratio was calculated by dividing the amplitude of eEPSCs by the amplitude of eIPSCs.
To measure the intrinsic excitability and rheobase using current-clamp recording, pipettes were filled with an intracellular solution containing: 125 mM K+-gluconate, 4 mM NaCl, 10 mM HEPES, 0.5 mM EGTA, 20 mM KCl, 4 mM Mg2+-ATP, 0.3 mM Na+-GTP and 10 mM Na2-phosphocreatine (290–300 mOsm, pH 7.2). To measure the firing capacity of LHLepr neurons, baseline firing was maintained and currents of increasing intensity (10-pA increments, duration of 500 ms) were injected up to 300 pA. Rheobase was determined as the minimal current step eliciting at least one action potential. All current-clamp recordings were performed in the presence of 5 µM NBQX (2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo(F)quinoxaline) and 50 µM picrotoxin to block synaptic transmission. Analyses were performed offline using Clampfit (Molecular Devices).
Intraperitoneal drug administration
For chemogenetic manipulations, CNO (Enzo Life Sciences) was dissolved in sterile distilled water. For the HFD intake experiments, mice were administered with CNO (2 mg per kg body weight, i.p.) or an equivalent volume of distilled water right before subjecting them to either HFD or Re-HFD. HFD intake was monitored for 2.5 h after CNO administration. For NC intake experiments, CNO (2 mg per kg body weight, i.p.) was administered either when NC was presented back after a 5-h food deprivation or during the basal state. Total NC intake levels were measured for 2.5 h after CNO administration. In the experiments analyzing pSTAT3 levels, leptin (Tocris Bioscience) was dissolved in sterile saline. Mice received an i.p. injection of leptin (1 mg per kg body weight) or saline 1 h before transcardial perfusion.
Intracranial drug injection and histology
On the day of the intracranial injection, an internal cannula (33-gauge, projecting 0.2 mm below the tip of the guide; Plastics One) connected to 1-ml syringes (Hamilton) via polyethylene (PE)−20 tubing was inserted into the guide. Leptin (1 µg per 0.5 µl per side, Tocris Bioscience) or saline was microinjected bilaterally into the LH when the NC was presented back after 5 h of food deprivation. Body weight and NC intake were monitored for 24 h after leptin administration.
For pegylated superactive mouse leptin antagonist (PESLAN, p.Asp23Leu/p.Leu39Ala/p.Asp40Ala/p.Phe41Ala mutant; Protein Laboratories Rehovot)70 experiment, sterile distilled water or PESLAN (1 or 2.5 µg per 0.5 µl per side) was bilaterally microinjected into the LH once a day during chronic NC or HFD exposure. Body weight changes of mice were recorded in home cages for 5 d of access to either NC or HFD. A high dose of PESLAN (2.5 µg per 0.5 µl per side) was microinjected bilaterally into the LH 5 min before the Re-HFD exposure in each binge-like eating cycle. HFD intake level was measured for 2.5 h.
For the chemogenetic experiment, CNO (1 mM, dissolved in artificial cerebrospinal fluid, 0.5 µl per side) was microinfused bilaterally into the vlPAG 5 min before Re-HFD exposure and the HFD intake level was monitored for 2.5 h.
Upon completion of experiments, mice were anesthetized and perfused with 4% PFA. Brains were extracted and further post-fixed in 4% PFA. Coronal sections (50 µm) were subsequently stained with 0.2% cresyl violet (Sigma-Aldrich) for the verification of cannula tip placements. Only mice with injection cannula tips located bilaterally in the LH or vlPAG were included in the data analysis.
Sample size and statistics
Samples sizes required for this study were initially estimated based on pilot studies, but no formal statistical tests were used to predetermine sample size. However, a power analysis was conducted to validate the sample size and the endpoint (significance level at 0.05 and power at 0.9).
If the viral expression was found outside this reference area or the viral transduction was weak in the LH (covering less than 50% of the total LH area), we excluded the mice from the final dataset, which was determined by two experimenters who were blinded to the experimental design. This exclusion happened in two wild-type mice (Extended Data Fig. 3n) due to the viral expression in VMH, one wild-type mouse (Extended Data Fig. 4a) due to off-target cannula implantation and one Lepr-Cre mouse (Fig. 5j) due to weak viral transduction.
We required the use of a group of animals with specific ages and a history of stress; however, within a group, we randomly chose animals for experiments. Animals used in this study were not selected based on any other prerequisite features other than general animal well-being (for example, normal grooming and social behavior, no obvious infections) for allocation into a particular experimental group.
Differences across more than two groups were analyzed with one-way or two-way ANOVA followed by Bonferroni or Fisher’s LSD post hoc tests for multiple comparisons. For comparisons between two groups, two-tailed t-tests (paired or unpaired) were used as described in the figure legends. The distribution of data was checked for normality and equal variance. P https://www.graphpad.com/).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.